Significance and bioactivity of resveratrol
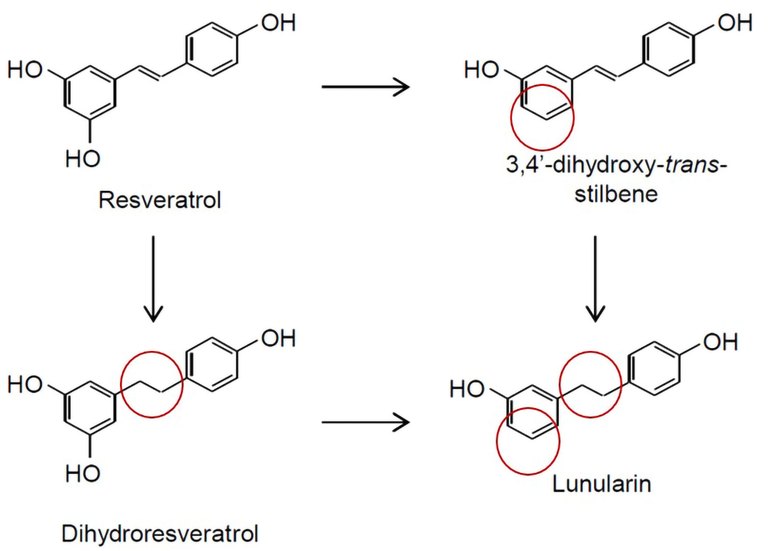
Resveratrol (3,5,4'-trihydroxy-trans-stilbene) is a secondary plant compound found in grapes, wine, various nuts, cocoa products and more. Several health-promoting effects of resveratrol are discussed. After oral intake, resveratrol is either metabolized directly by the human intestinal microbiota or it is absorbed by the intestinal mucosa and subsequently metabolized by endogenous enzymes. In addition to dihydroresveratrol, which has been known for some time, 3,4'-dihydroxy-trans-stilbene und lunularin were identified for the first time as microbial metabolites of resveratrol in a human study at the Max Rubner-Institut in a previous project [1] (Fig. 1). However, the quantitative relevance of these microbial metabolites in vivo and the microorganisms involved in this process were still poorly understood. Therefore, the aim of this project was to determine the relevance of resveratrol metabolites formed by the gut microbiota in humans.
For this purpose, a human intervention study was conducted with 104 healthy adults who ingested a single dose of 1 mg resveratrol per kg body weight. Subsequently, urine was collected over 48 h (0-24 h and 24-48 h). The amount of resveratrol and its metabolites excreted via urine was quantified by LC-MS/MS analysis. In the subjects, an average of almost two-thirds of the ingested amount of resveratrol was excreted via urine and was thus systemically available. The subjects showed large differences in the microbial metabolism of resveratrol. The metabolite dihydroresveratrol was determined in all volunteers with proportions of up to 80% in the renal excretion profile. Lunularin was detected in almost half of the volunteers, whereas 3,4'-dihydroxy-trans-stilbene was detected only to a minor extent. Thus, the microbial metabolites of resveratrol are quantitatively very relevant in humans.
Based on the different metabolite profiles, four metabolic phenotypes of resveratrol were described using a hierarchical cluster analysis and the volunteers were each assigned to one of these four phenotypes. In phenotype P1, resveratrol was highly metabolized. Phenotype P3, on the other hand, was characterized by relatively low metabolization of resveratrol. The extent of resveratrol metabolization in phenotype P2 was intermediate compared to phenotypes P1 (high) and P3 (low). The metabolite lunularin was formed in relevant amounts only in subjects of phenotype P4.
The question arises whether the large inter-individual differences in the metabolization of resveratrol have an influence on its biological effects, which could explain the partly contradictory research results on the health-promoting effects of resveratrol. This question should be addressed in further studies.
A special feature of this study was the characterization of the fecal microbiota of all 104 volunteers using 16S amplicon high-throughput sequencing. Enterotypes which were already described in the literature were also mapped in this group of subjects. However, a correlation of an enterotype or a bacterial taxon with a phenotype of the resveratrol metabolism was not found.
Another project objective was the identification of resveratrol- and dihydroresveratrol-metabolizing bacteria. For this purpose, 151 bacterial strains were isolated from a stool sample of a lunularin-producing volunteer under strictly anaerobic conditions. Of these, twelve Eggerthella lenta strains and one Adlercreutzia equolifaciens strain were identified that metabolize resveratrol to dihydroresveratrol. In addition, the type strains of Eggerthella lenta, Eggerthella sinensis, and Eggerthella timonensis were identified as dihydroresveratrol producers. These results highlight the importance of the genus Eggerthella for the microbial metabolism of resveratrol in humans.
As a further result of this study, two previously unknown, strictly anaerobic members of the bacterial family Eggerthellaceae were isolated from human fecal samples for the first time at the Max Rubner-Institut and taxonomically described: Rubneribacter badeniensis und Enteroscipio rubneri. These are strains of two previously unknown genera.
Scientific publication(s)
[1] Bode LM, Bunzel D, Huch M, Cho GS, Ruhland D, Bunzel M, Bub A, Franz CMAP, Kulling SE, 2013. In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota. American Journal of Clinincal Nutrition 97, 295-309.
https://pubmed.ncbi.nlm.nih.gov/23283496/
[2] Danylec N, Göbl A, Stoll DA, Hetzer B, Kulling SE, Huch M, 2018. Rubneribacter badeniensis gen. nov., sp. nov. and Enteroscipio rubneri gen. nov., sp. nov., new members of the Eggerthellaceae isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology 68, 1533-1540.
https://pubmed.ncbi.nlm.nih.gov/29537365/